-Search query
-Search result
Showing 1 - 50 of 728 items for (author: lo & hs)

EMDB-40260: 
CryoEM map of a de novo designed T=4 icosahedral nanocage hierarchically built from pseudosymmetric trimers; design Ico(T=4)-4
Method: single particle / : Borst AJ, Kibler RD, Lee S

EMDB-40249: 
CRISPR-Cas type III-D effector complex bound to a target RNA
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40248: 
CRISPR-Cas type III-D effector complex
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40250: 
CRISPR-Cas type III-D effector complex bound to a self-target RNA in the pre-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40251: 
CRISPR-Cas type III-D effector complex bound to self-target RNA in a post-cleavage state
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40276: 
CRISPR-Cas type III-D effector complex consensus map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40296: 
CRISPR-Cas type III-D effector complex local refinement map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40297: 
CRISPR-Cas type III-D effector complex bound to a target RNA local refinement map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40298: 
CRISPR-Cas type III-D effector complex bound to a target RNA consensus map
Method: single particle / : Schwartz EA, Taylor DW

EMDB-40267: 
CryoEM map of a T=1 off-target state of design Ico(T=4)-4
Method: single particle / : Borst AJ, Kibler RD, Lee S

EMDB-40268: 
CryoEM map of a de novo designed T=4 octahedral nanocage hierarchically built from pseudosymmetric trimers; design Oct(T=4)-3
Method: single particle / : Philomin A, Borst AJ, Kibler RD

EMDB-40269: 
CryoEM map of a T=1 off-target state of design Oct(T=4)-3
Method: single particle / : Philomin A, Borst AJ

EMDB-19477: 
Saccharomyces cerevisiae FAS type I
Method: single particle / : Mann D, Grininger M, Ludig D, Sachse C

EMDB-19489: 
Tobacco mosaic virus from scanning transmission electron microscopy at CSA=2.0 mrad
Method: helical / : Mann D, Filopoulou A, Sachse C
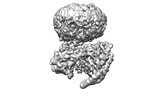
EMDB-42023: 
GPR3 Orphan G-coupled Protein Receptor in complex with Dominant Negative Gs.
Method: single particle / : Russell IC, Belousoff MJ, Sexton P

PDB-8u8f: 
GPR3 Orphan G-coupled Protein Receptor in complex with Dominant Negative Gs.
Method: single particle / : Russell IC, Belousoff MJ, Sexton P

EMDB-42480: 
Cryo-EM reconstruction of Staphylococcus aureus Oleate hydratase (OhyA) dimer with an ordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ

EMDB-42484: 
Cryo-EM reconstruction of Staphylococcus aureus oleate hydratase (OhyA) dimer with a disordered C-terminal membrane-association domain
Method: single particle / : Oldham ML, Qayyum MZ
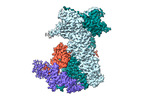
EMDB-18498: 
Cryo-EM structure of the benzo[a]pyrene-bound Hsp90-XAP2-AHR complex
Method: single particle / : Kwong HS, Grandvuillemin L, Sirounian S, Ancelin A, Lai-Kee-Him J, Carivenc C, Lancey C, Ragan TJ, Hesketh EL, Bourguet W, Gruszczyk J

EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD
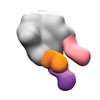
EMDB-40242: 
BG505 MD39 SOSIP in complex with Rh.NJ85 wk12 gp120GH, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40243: 
BG505 MD39 SOSIP in complex with Rh.NJ86 wk12 V1V3, C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB

EMDB-40244: 
BG505 MD39 SOSIP in complex with Rh.NJ76 wk12 C3V5, N611/FP, and base epitope polyclonal antibodies
Method: single particle / : Lee WH, Ozorowski G, Torres JL, Ward AB
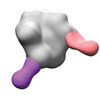
EMDB-40252: 
BG505 MD39 SOSIP in complex with Rh.NK04 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40254: 
BG505 MD39 SOSIP in complex with Rh.NJ75 wk12 gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40255: 
BG505 MD39 SOSIP in complex with Rh.NJ87 wk12 C3V5 and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40256: 
BG505 MD39 SOSIP in complex with Rh.NJ84 wk12 V1V3, gp120 glycan hole and base epitope polyclonal antibodies
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-40257: 
BG505 MD39 SOSIP in complex with Rh.NJ77 wk12 V1V3, C3V5, N611/FP and base epitope polyclonal
Method: single particle / : Lee WS, Ozorowski G, Torres JL, Ward AB

EMDB-16024: 
SARS-CoV-2 S protein in complex with pT1644 Fab
Method: single particle / : Stroeh L, Hansen G, Vollmer B, Krey T, Benecke T, Gruenewald K

EMDB-16026: 
SARS-CoV-2 S protein in complex with pT1696 Fab
Method: single particle / : Hansen G, Benecke T, Vollmer B, Gruenewald K, Krey T

EMDB-41066: 
Human VMAT2 in complex with tetrabenazine
Method: single particle / : Pidathala S, Dai Y, Lee CH

EMDB-41067: 
Human VMAT2 in complex with reserpine
Method: single particle / : Pidathala S, Dai Y, Lee CH

EMDB-41068: 
Human VMAT2 in complex with serotonin
Method: single particle / : Pidathala S, Dai Y, Lee CH

PDB-8t69: 
Human VMAT2 in complex with tetrabenazine
Method: single particle / : Pidathala S, Dai Y, Lee CH

PDB-8t6a: 
Human VMAT2 in complex with reserpine
Method: single particle / : Pidathala S, Dai Y, Lee CH

PDB-8t6b: 
Human VMAT2 in complex with serotonin
Method: single particle / : Pidathala S, Dai Y, Lee CH

EMDB-18150: 
Cryo-EM map of the Candida albicans 80S ribosome in complex with cephaeline
Method: single particle / : Kolosova O, Zgadzay Y, Stetsenko A, Atamas A, Jenner L, Guskov A, Yusupov M

EMDB-18151: 
Focused map on the body of the SSU of the Candida albicans 80S ribosome in complex with cephaeline
Method: single particle / : Kolosova O, Zgadzay Y, Stetsenko A, Atamas A, Jenner L, Guskov A, Yusupov M

EMDB-18155: 
The cryo-EM map of the C. albicans ribosome focused on the head of the SSU
Method: single particle / : Kolosova O, Zgadzay Y, Stetsenko A, Atamas A, Jenner L, Guskov A, Yusupov M

EMDB-17015: 
Cryo-EM map of the focused refinement of the subfamily III haloalkane dehalogenase from Haloferax mediterranei dimer forming hexameric assembly.
Method: single particle / : Polak M, Novacek J, Chmelova K, Marek M

PDB-8ooh: 
Cryo-EM map of the focused refinement of the subfamily III haloalkane dehalogenase from Haloferax mediterranei dimer forming hexameric assembly.
Method: single particle / : Polak M, Novacek J, Chmelova K, Marek M
EMDB-28617: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB
Method: single particle / : Pletnev S, Kwong P

EMDB-28618: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB
Method: single particle / : Pletnev S, Kwong P

EMDB-28619: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB
Method: single particle / : Pletnev S, Kwong P

PDB-8euu: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB
Method: single particle / : Pletnev S, Kwong P

PDB-8euv: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB
Method: single particle / : Pletnev S, Kwong P

PDB-8euw: 
Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB
Method: single particle / : Pletnev S, Kwong P

EMDB-28115: 
Western Equine Encephalitis Virus-Like Particle in Complex with SKW19 Fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong P

EMDB-28116: 
Western Equine Encephalitis Virus-Like Particle in Complex with SKW24 Fab
Method: single particle / : Pletnev S, Tsybovsky Y, Verardi R, Roedeger M, Kwong PD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
